

Preview of Topics at HPLC 2016, IV
From the Research Lab to Routine Characterization
By Alain Beck
Senior Director of the Physico-Chemistry Department
Centre d’Immunologie Pierre Fabre (CIPF), Saint-Julien-en-Genevois, France
and Arnaud Delobel
Scientific Manager, Biologics / R&D Team Manager
Quality Assistance s.a., Belgium
This is the fourth in a series of four articles exploring topics that will be addressed at the HPLC 2016 conference in San Francisco, from June 19 to 24.
Monoclonal antibodies (mAbs) are highly complex tetrameric glycoproteins that require extensive analytical and structural characterization to become drug candidates. This is also true for antibody–drug conjugates (ADCs). These immunoconjugates are based on highly cytotoxic small-molecule drugs covalently attached via conditionally stable linkers to mAbs and are among the most promising next-generation empowered biologics for cancer treatment. ADCs are more complex than naked mAbs, because the heterogeneity of the conjugates adds to the inherent microvariability of the biomolecules. The development and optimization of ADCs rely on improving their analytical and bioanalytical characterization by assessing several critical quality attributes, namely the distribution and position of the drug, the amount of naked antibody, the average drug-to-antibody ratio (DAR), and the proportions of residual small-molecule drug and drug-linker) (1).
As a result of advances in multilevel (top, middle, bottom) state-of-the-art mass spectrometry (MS) methods, including native MS, ion mobility MS (2), capillary electrophoresis–electrospray ionization–MS (3,4), two-dimensional liquid chromatography–MS (2D LC–MS) (5), extended bottom-up (6), and top-down sequencing (7), combined with chromatographic and electrophoretic techniques (8) very precise characterization of biotherapeutics is now possible. Until recently, however, these techniques were considered suitable only for research use. With the advent of robust and user-friendly solutions (both hardware and software), these techniques are now amenable for routine use. Examples of their application to the characterization of mAbs and ADCs are discussed below.
During their biosynthesis or during their shelf life, mAbs can undergo many modifications, such as glycosylation, oxidation, deamidation, and C-terminal lysine clipping, to name a few. In most cases, these variants cannot be easily identified at the intact antibody level, because of limitations in chromatographic separation and MS resolution.
A middle-up approach using IdeS digestion (to yield Fab, Fc/2, and light-chain fragments) combined with a super macroporous reversed-phase column enables quick and efficient characterization of mAb variants (9). The relatively low molecular mass of the subunits (~25 kDa) allows accurate mass determination by high-resolution MS as well as top-down sequencing by electron transfer dissociation (ETD). Using the same sample preparation, the glycoforms can also be separated and characterized using an approach such as hydrophilic interaction chromatography (HILIC) with MS detection (10). An example of this approach is presented in Figure 1 for adalimumab.
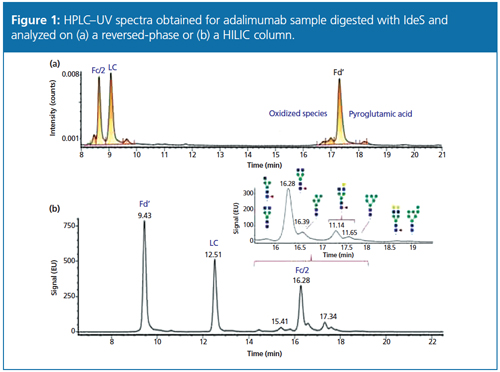
Figure 1: HPLC–UV spectra obtained for adalimumab sample digested with IdeS and analyzed on (a) a reversed-phase or (b) a HILIC column.
The number of cytotoxic molecules attached to an antibody (the drug-to-antibody ratio, or DAR) is a critical quality attribute of an ADC. It can be determined by UV spectrophotometry or hydrophobic interaction chromatography (HIC) with UV detection (11–13), but these methods are not universal and have some drawbacks. Mass spectrometry can be a universal tool to determine the DAR value, whatever the coupling chemistry or the nature of the drugs.
After N-deglycosylation, the ADC is desalted online using reversed-phase high performance LC (HPLC) or size-exclusion chromatography (SEC) (14) and detected by high-resolution electrospray ionization MS. The DAR can be automatically calculated by software after deconvolution of the multicharged electrospray spectrum. When working with cysteine-linked ADCs, native conditions must be used to avoid disruption of noncovalent interactions. In these methods, native SEC–MS is commonly used, which requires optimization of mobile phases and MS conditions.
Peptide mapping with MS detection is a common methodology for protein characterization. It can be used for the confirmation of the primary sequence, the quantification of post-translational modifications (PTMs), and the study of disulfide-bond scrambling. Thanks to commercially available software packages dedicated to biopharmaceutical analysis, the analysis of these MS data can be fully automated.
Applied to ADC characterization, peptide mapping is also a valuable tool to localize conjugation sites and determine site occupancy. ETD fragmentation can be used to localize conjugation sites for lysine-conjugated ADCs on peptides containing several lysine residues.
Two-dimensional LC with MS detection is widely used for the identification of proteins from complex proteome samples in many laboratories. Robust 2D–HPLC and 2D-UHPLC systems are now commercially available, thus enabling the routine analysis of biopharmaceuticals with this technology.
Two-dimensional LC can be used to hyphenate MS-incompatible chromatographic separations to mass spectrometry detection: After a first dimension using mobile phases containing nonvolatile salts, the peak of interest is sent to a second dimension consisting of a reversed-phase column to desalt the sample and separate potentially coeluted species. This methodology (heart-cutting 2D LC–MS) can be routinely applied to identify the different species detected in size-exclusion and ion-exchange chromatography of monoclonal antibodies.
Another main application of 2D LC–MS is the rapid on-line structural elucidation of species observed in HIC distribution profiles of cysteine-conjugated ADCs (15,16). The identification of the different species is required to be able to determine the DAR value based on the HIC–UV profile as well as for the detection and the quantification of small-molecule drugs (17). The application of this methodology to brentuximab vedotin is presented in Figure 2.
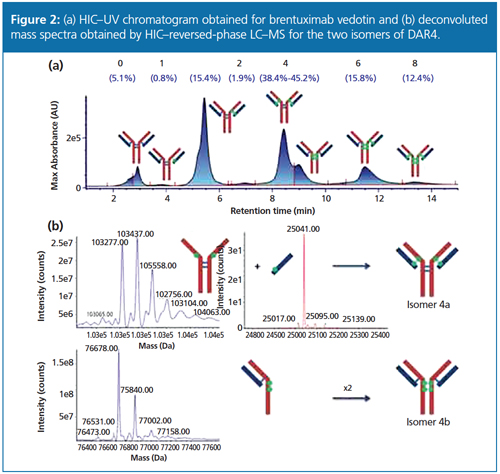
Figure 2: (a) HIC–UV chromatogram obtained for brentuximab vedotin and (b) deconvoluted mass spectra obtained by
HIC–reversed-phase LC–MS for the two isomers of DAR4.
All these new techniques and case studies will be discussed at the HPLC 2016 meeting in San Francisco in June.