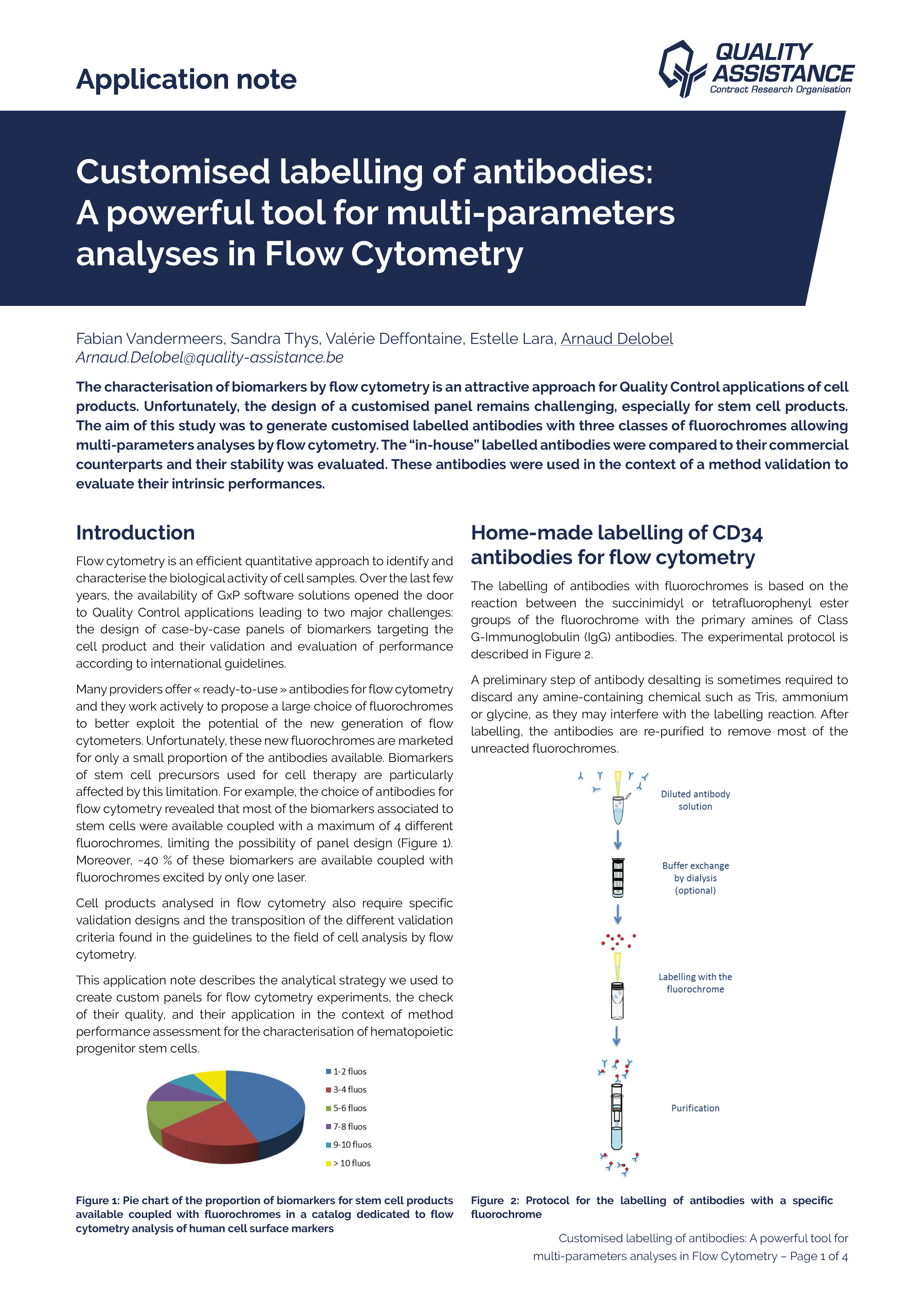
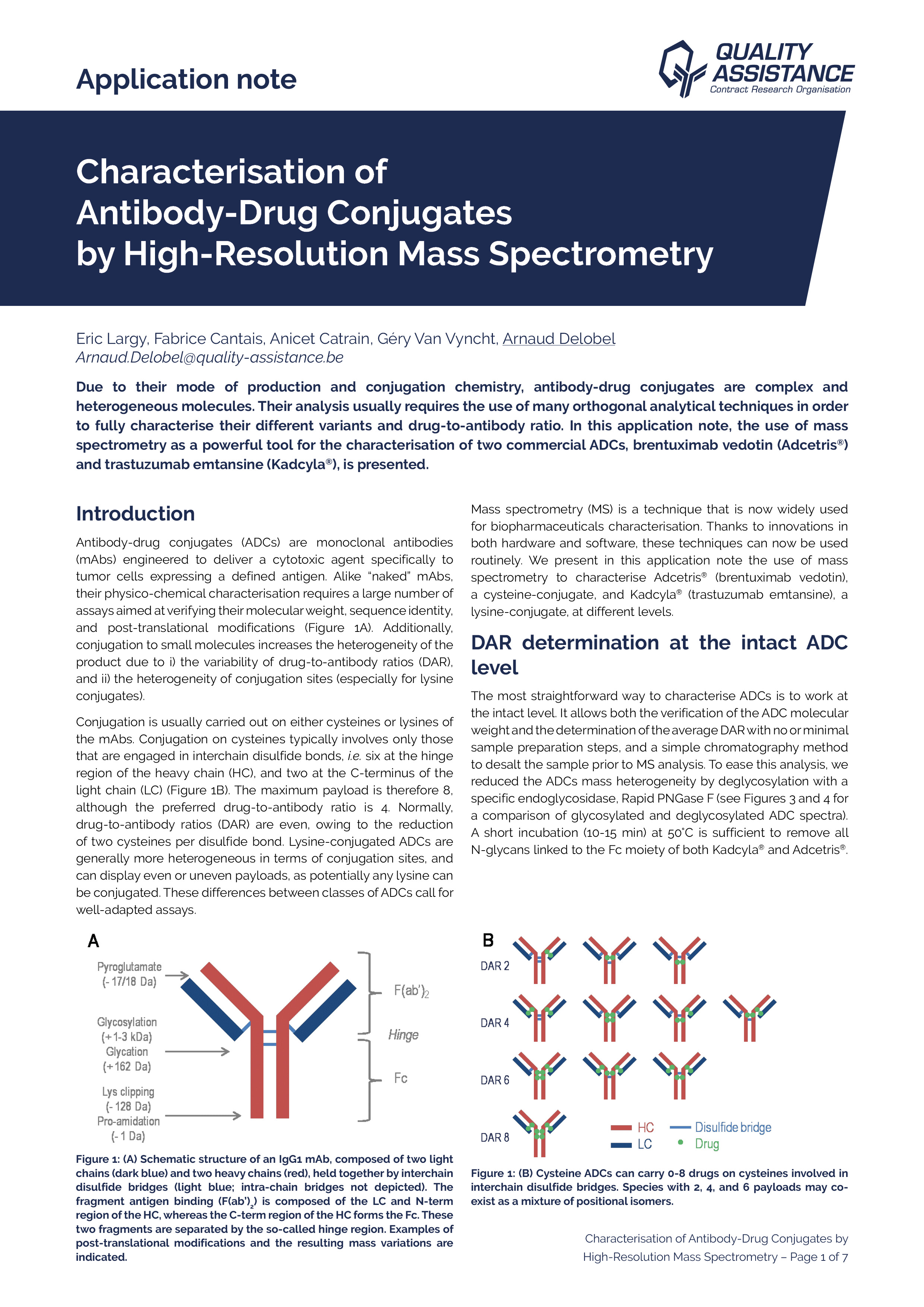
Due to their mode of production, therapeutic mAbs present diverse and heterogeneous glycosylations. Their analysis may require the use of orthogonal techniques in order to fully characterize their nature and location. In this application note, the use of mass spectrometry as a powerful tool for Adalimumab and Cetuximab glycosylation characterization is presented.
Using a comprehensive workflow combining several analytical methods at the subunit, peptide, and released glycan levels, we extensively characterised the N-glycosylation of adalimumab and cetuximab with great consistency.
Profiling of the N-glycans was performed within a day using the RapiFluor-MS labelling kit (Waters), and is fully consistent with results from the literature. The Neu5Ac and Neu5Gc sialylation was quantified by straightforward acid-induced release and DMBlabelling, starting from small amounts of mAbs. The results are consistent with the N-glycosylation profiles determined previously.
Profiling of each of the N-glycosylation sites was performed by widepore HILIC/MS, after triple enzymatic digestion, yielding short peptides and a complete sequence coverage. This stationary phase allows a complete resolution of glycosylated peptides vs. aglycosylated peptides, and MSE data provides a confirmation of
the presence of glycans. We evidenced significant differences in the glycosylation of the two cetuximab sites, the overall profile being consistent with what was found by released glycans analysis.
Analysis of mAbs subunits, generated by IdeS enzyme digestion, followed by an in situ reduction, confirmed the glycosylation profile and the localisation of the glycosylation sites, with a much easier and faster sample preparation protocol than for the peptide mapping approach. It is also a way to confirm the molecular weight of these subunits, which may be challenging because of the important heterogeneity of glycosylation and other PTMs.
This comprehensive workflow is fully applicable to potentially any mAb or ADC, using small quantities of sample, and in a reduced amount of time. Other analytical methods are available at Quality Assistance to complete the study of therapeutic mAbs glycosylation, such as relative quantification of the fucosylation, sialylation profiling by mixed-mode chromatography, and MALDI TOF profiling.